Contents
Note: This tutorial assumes that the computer to be used already has a functioning installation of the latest version of the Starlink software suite (the latest release is available for download here).
This tutorial aims to go through some of the many analysis tools within Starlink.
Get the data:
Download the tarball from HERE. This contains
- An archival JCMT CO 3-2 HARP reduced map:
jcmth20061220_00044_01_reduced001_nit_000.fits - A reduced 850um SCUBA-2 map of G34.3, as produced in the Basic SCUBA-2 DR tutorial.
scuba2_850.sdf - A TST catalogue file showing the JCMT state information from a SCUBA-2 raw obsrvation:
jcmtstate_s20120501_00068.TST
Start Starlink
If using BASH/ZSH:
export STARLINK_DIR=/path/to/starlink/directory/star-2017A source $STARLINK_DIR/etc/profile
If using TCSH/CSH:
setenv STARLINK_DIR /path/to/starlink/directory/star-2017A source $STARLINK_DIR/etc/login source $STARLINK_DIR/etc/profile
Getting started with your data
Task: Convert FITS cube to Radio Velocity SDF
Take a HARP cube downloaded from the JCMT Science Archive in FITS format, convert it back to a .sdf NDF file, and then change the barycentric frequency axis into a radio velocity axis with the standard of rest set to LSRK.
Click to show worked solutionWorked Example
The first file we are going to look at is an archival HARP map jcmth20061220_00044_01_reduced001_nit_000.fits
This is a 2006 reduced cube containing a CO 3-2 HARP observation of Ros 6.
The first thing we will do is convert it from FITS format into NDF — the Starlink data format. To do this we need to inialise the convert package:
$convert CONVERT commands are now available -- (Version 1.8) Defaults for automatic NDF conversion are set. Type conhelp for help on CONVERT commands. Type "showme sun55" to browse the hypertext documentation.
We then run the command fits2ndf:
$fits2ndf jcmth20061220_00044_01_reduced001_nit_000.fits co32_cube.sdf
JCMT archive data uses Barycentric Frequency as its frequency coordinate system; use ndftrace to examine this. First of all you will need to initialise the package by typing kappa. Then, we will use ndftrace to see what is in this file.
(Throughout this tutorial we often will show you the output of the commands. To make it clear what is output vs what is input this tutorial puts a $ at the start of the command you need to type, indicating the shell prompt.)
$ ndftrace co32_cube.sdf
NDF structure
/export/data/sgraves/sd/tutorial-data/Starlink_Analysis/co32_cube:
Title: Ros6
Label: TA* corrected antenna temperature
Units: K
Shape:
No. of dimensions: 3
Dimension size(s): 119 x 118 x 1880
Pixel bounds : -57:61, -58:59, -940:939
Total pixels : 26398960
Data Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
Variance Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
World Co-ordinate Systems:
Number of co-ordinate Frames: 5
Current co-ordinate Frame (Frame 5):
Frame title : "3-d compound coordinate system"
Domain : SKY-DSBSPECTRUM
First pixel centre : 6:35:01.2, 4:06:50, 346.2167
Axis 1:
Label : Right ascension
Units : hh:mm:ss.s
Nominal Pixel scale: 5.99999 arc-sec
Axis 2:
Label : Declination
Units : ddd:mm:ss
Nominal Pixel scale: 5.99999 arc-sec
Axis 3:
Label : Frequency (USB)
Units : GHz
Nominal Pixel scale: 0.0004882367 GHz
Extensions:
FITS <_CHAR*80>
SMURF
PROVENANCE
History Component:
Created : 2014 Nov 20 09:22:05
No. records: 20
Last update: 2014 Nov 20 09:23:59 (FITSMOD (KAPPA 2.2))
Update mode: NORMAL
This shows you the label, units, number and shape of the dimensions, some of the coordinate values, and also shows you the extensions in the NDF file.
You can see that the third axis is Frequency, in units of GHz. We can check what standard of rest it is using the wcsattrib command:
$ wcsattrib co32_cube.sdf mode=get name=StdofRest Barycentric
We are going to change the third axis to be in Radio Velocity, and we are also going to change to the LSRK standard of rest, again using the wcsattrib but this time in mode=set. To see the list of WCS information you can see/alter with wcsattrib, refer to the KAPPA user note (SUN/95) at www.starlink.ac.uk/cgi-bin/htxserver/sun95.htx/sun95.html?xref_ap_frmatt
# Change to radio velocity (vrad) and the LSRK std of rest. wcsattrib ndf=co32_cube.sdf mode=set name=system newval=vrad wcsattrib ndf=co32_cube.sdf mode=set name=StdofRest newval=LSRK
If we look at again at our file with ndftrace, you can see the changes to the 3rd axis:
$ ndftrace co32_cube.sdf
NDF structure
/export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/co
32cube:
Title: Ros6
Label: TA* corrected antenna temperature
Units: K
Shape:
No. of dimensions: 3
Dimension size(s): 119 x 118 x 1880
Pixel bounds : -57:61, -58:59, -940:939
Total pixels : 26398960
Data Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
Variance Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
World Co-ordinate Systems:
Number of co-ordinate Frames: 5
Current co-ordinate Frame (Frame 5):
Frame title : "3-d compound coordinate system"
Domain : SKY-DSBSPECTRUM
First pixel centre : 6:35:01.2, 4:06:50, -381.1352
Axis 1:
Label : Right ascension
Units : hh:mm:ss.s
Nominal Pixel scale: 5.99999 arc-sec
Axis 2:
Label : Declination
Units : ddd:mm:ss
Nominal Pixel scale: 5.99999 arc-sec
Axis 3:
Label : Radio velocity (USB)
Units : km/s
Nominal Pixel scale: 0.4233065 km/s
Extensions:
FITS <_CHAR*80>
SMURF
PROVENANCE
History Component:
Created : 2014 Nov 20 09:22:05
No. records: 23
Last update: 2018 Jan 25 20:03:17 (NDFCOPY (KAPPA 2.5-5))
Update mode: NORMAL
You can see the 3rd axis in now in units of km/s, with a pixel scale 0.423 km/s per channel.
Task: Examining what is in your file
Looking at the cube file you produced above, identify the molecule and transition, the original date of the observation and what. Find out if the autogrid option was used in the original makecube command that gridded the data, and when that command was run. Find out all the original observations and files that were included in the file. Finally, see what extensions are present in your data file.
Click to show worked solutionWe’ve already used KAPPA’s wcsattrib and ndftrace above to look at what is in your file. There are several other commands that can be helpful.
- fitslist This command shows you the contents of the FITS extension of the NDF. This is where we store information about the observation the file was produced by (including integration time, scan pattern, tracking system) , the telescope information, and the environmental information.
$ fitslist ../Starlink_Analysis/co32_cube.sdf
-
SIMPLE = T / file does conform to FITS standard BITPIX = -32 / number of bits per data pixel NAXIS = 3 / number of data axes NAXIS1 = 119 / length of data axis 1 NAXIS2 = 118 / length of data axis 2 NAXIS3 = 1880 / length of data axis 3 EXTEND = T / FITS dataset may contain extensions COMMENT FITS (Flexible Image Transport System) format is defined in 'Astronomy COMMENT and Astrophysics', volume 376, page 359; bibcode: 2001A&A...376..359H LBOUND1 = -57 / Pixel origin along axis 1 LBOUND2 = -58 / Pixel origin along axis 2 LBOUND3 = -940 / Pixel origin along axis 3 LABEL = 'T%+%+A%^50+%NUMTILES) INSTREAM= 'JCMT ' / Source of input stream DPDATE = '2014-11-20T09:23:54' / Data processing date DPRCINST= 'jac-92883' / Data processing recipe instance ID / Provenance: PRVCNT = 6 / Number of parents PRV1 = 'a20061220_00044_01_0001' / Name of the 1st parent PRV2 = 'a20061220_00044_01_0002' / Name of the 2nd parent PRV3 = 'a20061220_00044_01_0003' / Name of the 3rd parent PRV4 = 'oractempuMu_kY' / Name of the 4th parent PRV5 = 'oractempEjA5Sw' / Name of the 5th parent PRV6 = 'oractempSgkQoo' / Name of the 6th parent OBSCNT = 1 / Number of root-ancestor headers OBS1 = 'acsis_44_20061220T095454_1' / Name of the 1st root ancestor FILEID = 'jcmth20061220_00044_01_reduced001_nit_000' / Filename minus extension CHECKSUM= '0a7F3T6D0Z6D0Z6D' / HDU checksum updated 2014-11-20T09:24:00 DATASUM = '1474200105' / data unit checksum updated 2014-11-20T09:24:00 ENDYou can also get the information from a specific header with ‘fitsval’. E.g. to find the value of the instrume keyword
$ fitsval co32_cube.sdf keyword=instrume HARP
- hislist will show you the Starlink commands that were carried out on the file. This is very useful for checking that you have done what you thought you did to a file, or when you come back to work again on files after a long pause.
$ hislist co32_cube.sdf History listing for NDF structure /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/co32_cube: History structure created 2014 Nov 20 09:22:05.443 1: 2014 Nov 20 09:22:05.538 - MAKECUBE (SMURF V1.6.0) Parameters: ALIGNSYS=FALSE AUTOGRID=TRUE BADMASK='and' CROTA=0 DETECTORS=! FBL=[1.7235973391403,0.071799010054101] FBR=[1.7201560002942,0.071799006183432] FLBND=[1.7201409775556,0.07178446001926,-381.34682629178] FUBND=[1.7236123539655,0.07521704716548,414.46932659916] FTL=[1.7235977665515,0.075202393861675] FTR=[1.7201555650017,0.075202389806852] GENVAR='tsys' IN=@^oractempXdGhiP INWEIGHT=TRUE JSATILES=FALSE LBND=[-57,-58] LBOUND=[-57,-58,-940] NPOLBIN=1 NTILE=1 OUT=@ga20061220_44_1_cube OUTCAT=! PIXREF=[60.461,60.045] PIXSIZE=6 POSERRFATAL=FALSE REF=! REFLAT='4:12:43.9' REFLON='6:34:37.72' REFPIX1=60.461 REFPIX2=60.045 SPARSE=FALSE SPECBOUNDS='-381.347,414.469' SPECUNION=TRUE SPREAD='NEAREST' SYSTEM='tracking' TILEBORDER=0 TILEDIMS=-256 TRIM=FALSE TRIMTILES=TRUE UBND=[61,59] UBOUND=[61,59,939] USEDETPOS=FALSE WEIGHTS=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/smurf/smurf_mon Group: IN="a20061220_00044_00_bl001, a20061220_00044_00_bl002, a20061220_00044_00_bl003" 2: 2014 Nov 20 09:22:22.324 - FITSMOD (KAPPA 2.2) Parameters: COMMENT='$' EDIT='update' EXISTS=TRUE KEYWORD='DATE-OBS' MODE='Interface' NDF=@ga20061220_44_1_cube001 POSITION=! READONLY=FALSE STRING=TRUE VALUE='2006-12-20T09:54:54' MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 3: 2014 Nov 20 09:22:22.374 - FITSMOD (KAPPA 2.2) Parameters: COMMENT='$' EDIT='update' EXISTS=TRUE KEYWORD='DATE-END' MODE='Interface' NDF=@ga20061220_44_1_cube001 POSITION=! READONLY=FALSE STRING=TRUE VALUE='2006-12-20T10:33:33' MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 4: 2014 Nov 20 09:22:22.884 - ORAC-DR - _CREATE_CUBE_GROUP_ (29bb4c) Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_cube001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 5: 2014 Nov 20 09:22:34.018 - SUB (KAPPA 2.2) Parameters: IN1=@ga20061220_44_1_cube001 IN2=@oractempS28Rf4 OUT=@ga20061220_44_1_bl001 TITLE=! MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/kappa_mon 6: 2014 Nov 20 09:22:34.759 - SETVAR (KAPPA 2.2) Parameters: COMP='VARIANCE' FROM=@ga20061220_44_1_cube001 NDF=@ga20061220_44_1_bl001 MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 7: 2014 Nov 20 09:22:35.894 - ORAC-DR - _REMOVE_BASELINE_THROUGH_SMOOTHING_ ... Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_bl001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 8: 2014 Nov 20 09:22:36.027 - SETBB (KAPPA 2.2) Parameters: AND=FALSE BB='255' NDF=@ga20061220_44_1_bl001 OR=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 9: 2014 Nov 20 09:22:36.328 - NDFCOPY (KAPPA 2.2) Parameters: COMP='DATA' EXTEN=FALSE IN=@ga20061220_44_1_bl001 LIKE=! OUT=@ga20061220_44_1_reduced001 TITLE=! TRIM=FALSE TRIMBAD=FALSE TRIMWCS=TRUE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 10: 2014 Nov 20 09:22:38.077 - ORAC-DR - _TAG_AS_REDUCED_PRODUCT_ (29bb4c) Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_reduced001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 11: 2014 Nov 20 09:22:38.215 - SETBB (KAPPA 2.2) Parameters: AND=FALSE BB='0' NDF=@ga20061220_44_1_reduced001 OR=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 12: 2014 Nov 20 09:22:41.082 - SETBB (KAPPA 2.2) Parameters: AND=FALSE BB='255' NDF=@ga20061220_44_1_reduced001 OR=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 13: 2014 Nov 20 09:23:43.241 - ORAC-DR - _CREATE_MOMENTS_MAPS_THROUGH_SMOOTH... Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_reduced001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 14: 2014 Nov 20 09:23:44.445 - SETBB (KAPPA 2.2) Parameters: AND=FALSE BB='0' NDF=@ga20061220_44_1_reduced001 OR=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 15: 2014 Nov 20 09:23:44.847 - SETBB (KAPPA 2.2) Parameters: AND=FALSE BB='255' NDF=@ga20061220_44_1_reduced001 OR=FALSE MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/ndfpack_mon 16: 2014 Nov 20 09:23:48.025 - ORAC-DR - _CREATE_MOMENTS_MAPS_THROUGH_SMOOTH... Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_reduced001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 17: 2014 Nov 20 09:23:49.451 - ORAC-DR - _CREATE_NOISE_MAP_ (29bb4c) Astro::FITS::Header::NDF - write FITS header to file /export/data/jsa_proc/scratch/000/000092/000092883/2014 -11 -20_08-49-03/ga20061220_44_1_reduced001 Arguments: -nodisplay -log hs -verbose -recsuffix ADV,CADC -batch -loop file -file /tmp/7hzdep6Ydp -recpars recpars-M06BH23B.ini Software: /net/kamaka/export/data/stardev-stable/bin/oracdr/src/b in/ oracdr 18: 2014 Nov 20 09:23:59.727 - jsawrapdr Arguments: --debugxfer --outdir=/export/data/jsa_proc/scratch/000/000092/000092883/2014-11-2 8-49-03 --inputs=/net/kamaka/export/data/jsa_proc/input/000/000092/000092883/ ut_files_job.lis --id=jac-92883 --mode=night --cleanup=cadc --drparameters=-recpars recpars-M06BH23B.ini -persist --transdir=/net/kamaka/export/data/jsa_proc/output/000/000092/0000928 Software: /net/kamaka/export/data/stardev-stable/Perl/bin/jsawrapdr 19: 2014 Nov 20 09:23:59.772 - WCSATTRIB (KAPPA 2.2) Parameters: MODE='MSet' NDF=@ga20061220_44_1_reduced001.sdf REMAP=TRUE SETTING='System(1)=FK5,System(3)=FREQ,StdOfRest=BARY' MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/wcsattrib 20: 2014 Nov 20 09:23:59.835 - FITSMOD (KAPPA 2.2) Parameters: EXISTS=TRUE MODE='File' NDF=@ga20061220_44_1_reduced001.sdf READONLY=FALSE TABLE=@/tmp/PjQSB6jkf9 MSG_FILTER=! QUIET=! Software: /net/kamaka/export/data/stardev-stable/bin/kappa/fitsmod 21: 2018 Jan 25 22:26:19.487 - WCSATTRIB (KAPPA 2.5-5) Parameters: MODE='set' NAME='system' NDF=@co32_cube.sdf NEWVAL='vrad' REMAP=TRUE STATE=FALSE VALUE='Compound' MSG_FILTER=! QUIET=! Software: /star/bin/kappa/wcsattrib 22: 2018 Jan 25 22:26:20.955 - WCSATTRIB (KAPPA 2.5-5) Parameters: MODE='set' NAME='StdofRest' NDF=@co32_cube.sdf NEWVAL='LSRK' REMAP=TRUE STATE=TRUE VALUE='Barycentric' MSG_FILTER=! QUIET=! Software: /star/bin/kappa/wcsattribCan you see the 2 wcsattrib commands you just ran on the file?
- provshow lets you see the NDF files that were used to create the file you are exmaining. Its mostly commonly used to check which observations you included in a coadded file.
$ provshow co32_cube.sdf 0: /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/co32_cube Parents: 1,2,3,6,5,4 Date: 2014-11-20 09:23:32 Creator: KAPPA:NDFCOPY 1: a20061220_00044_01_0001.sdf Parents: Date: Creator: More: OBSIDSS=acsis_44_20061220T095454_1 2: a20061220_00044_01_0002.sdf Parents: Date: Creator: More: OBSIDSS=acsis_44_20061220T095454_1 3: a20061220_00044_01_0003.sdf Parents: Date: Creator: More: OBSIDSS=acsis_44_20061220T095454_1 4: ./oractempSgkQoo.sdf Parents: Date: Creator: 5: ./oractempEjA5Sw.sdf Parents: Date: Creator: 6: ./oractempuMu_kY.sdf Parents: Date: Creator:
Can you see which observation was included. How many raw files went into this cube?
- hdstrace shows you the entire structure of the file. This lets you see any extensions included in the data.
$ hdstrace co32_cube.sdf CO32_CUBE DATA_ARRAY {structure} ORIGIN(3) <_INTEGER> -57,-58,-940 DATA(119,118,1880) <_REAL> *,*,*,-3.457747,1.411959,-2.120131,0.4018285,-1.767746,-0.7077882,1.616902,0.9329623,-1.961738,0.8053908,1.151465,6.122407,2.263416,-3.084188,-0.8098075,0.4601... ... 0.4417757,3.771597,-0.383148,1.626338,-0.6567043,-2.779671,2.554621,-1.354576,1.24353,-2.09212,-0.320193,1.203157,-2.560659,0.1859031,*,-0.1394619,-4.89816... HISTORY {structure} CREATED <_CHAR*24> '2014-NOV-20 09:22:05.443' UPDATE_MODE <_CHAR*8> 'NORMAL' CURRENT_RECORD <_INTEGER> 22 RECORDS(25) {array of structures} Contents of RECORDS(1) DATE <_CHAR*24> '2014-NOV-20 09:22:05.538' COMMAND <_CHAR*30> 'MAKECUBE (SMURF V1.6.0)' USER <_CHAR*7> 'jsaproc' HOST <_CHAR*6> 'ikaika' DATASET <_CHAR*70> '/export/data/jsa_proc/scratch/000/000092/000092883/2014-11-20_08-49...' TEXT(18) <_CHAR*72> 'Parameters: ALIGNSYS=FALSE AUTOGRID=TRUE BADMASK='and' CROTA=0',' DETECTORS=! FBL=[1.7235973391403,0.071799010054101]',' FBR=[1.7201560002942,0.0717990... ... ' USEDETPOS=FALSE WE...','Software: /net/kamaka/export/data/stardev-stable/bin/smurf/smurf_mon','Group: IN="a20061220_00044_00_bl001, a20061220_00044_... MORE {structure} FITS(273) <_CHAR*80> 'SIMPLE = T / file does conform to FITS standard','BITPIX = -32 / number of bits per data pixel','NAXIS = ... 'FILEID = 'jcmth20061220...','','CHECKSUM= '0a7F3T6D0Z6D0Z6D' / HDU checksum updated 2014-11-20T09:24:00','DATASUM = '1474200105' / data unit ch... SMURF {structure} EXP_TIME {structure} DATA_ARRAY {structure} ORIGIN(2) <_INTEGER> -57,-58 DATA(119,118) <_REAL> *,*,*,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05... ... 2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2.05,2... TITLE <_CHAR*4> 'Ros6' LABEL <_CHAR*19> 'Total exposure time' UNITS <_CHAR*1> 's' WCS {structure} DATA(178) <_CHAR*32> ' Begin FrameSet',' Nframe = 5',' Currnt = 5',' Lnk2 = 1',' Lnk3 = 1',' Lnk4 = 1',' Lnk5 = 1',' Frm1 =',' Begin Frame',' Naxes = 2',' Domain = "GRID"'... ' Nout = 2',' Invert = 0',' IsA Mapping',' UntRd = 1',' PlrLg = 1.72189238902213',' End SphMap',' End CmpMap',' End CmpMap',' End CmpMap',' End CmpMap... EFF_TIME {structure} DATA_ARRAY {structure} ORIGIN(2) <_INTEGER> -57,-58 DATA(119,118) <_REAL> *,*,*,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.38... ... 0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,0.3804878,*,0.3804878,0.76... TITLE <_CHAR*4> 'Ros6' LABEL <_CHAR*26> 'Effective integration time' UNITS <_CHAR*1> 's' WCS {structure} DATA(178) <_CHAR*32> ' Begin FrameSet',' Nframe = 5',' Currnt = 5',' Lnk2 = 1',' Lnk3 = 1',' Lnk4 = 1',' Lnk5 = 1',' Frm1 =',' Begin Frame',' Naxes = 2',' Domain = "GRID"'... ' Nout = 2',' Invert = 0',' IsA Mapping',' UntRd = 1',' PlrLg = 1.72189238902213',' End SphMap',' End CmpMap',' End CmpMap',' End CmpMap',' End CmpMap... TSYS {structure} DATA_ARRAY {structure} ORIGIN(2) <_INTEGER> -57,-58 DATA(119,118) <_REAL> *,*,*,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,388.4387,... ... 371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,371.6322,*,371.6322,... TITLE <_CHAR*4> 'Ros6' LABEL <_CHAR*28> 'Effective system temperature' UNITS <_CHAR*1> 'K' WCS {structure} DATA(178) <_CHAR*32> ' Begin FrameSet',' Nframe = 5',' Currnt = 5',' Lnk2 = 1',' Lnk3 = 1',' Lnk4 = 1',' Lnk5 = 1',' Frm1 =',' Begin Frame',' Naxes = 2',' Domain = "GRID"'... ' Nout = 2',' Invert = 0',' IsA Mapping',' UntRd = 1',' PlrLg = 1.72189238902213',' End SphMap',' End CmpMap',' End CmpMap',' End CmpMap',' End CmpMap... PROVENANCE {structure} DATA(345) <_INTEGER> 2,7,1598506062,1280070990,825242112,825306420,540029485,842676528,842218035,1095434240,977358928,1128678478,5853263,1481880154,-1,6,1,2,3,6,5,4,808464993,84... ... 1431199559,19532,-16777216,16777215,771751936,1634889519,1835365475,1968010608,777612127,6710387,1598506062,1280070990,1145962496,1431199559,19532,-1677... TITLE <_CHAR*4> 'Ros6' LABEL <_CHAR*57> 'T%+%+A%^50+%<20+*%+ corrected antenna temperature' UNITS <_CHAR*1> 'K' VARIANCE {structure} DATA(119,118,1880) <_REAL> *,*,*,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,4.915271,... ... 4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,4.499031,*,4.499031,... ORIGIN(3) <_INTEGER> -57,-58,-940 WCS {structure} DATA(274) <_CHAR*32> ' Begin FrameSet',' Nframe = 5',' Currnt = 5',' Lnk2 = 1',' Lnk3 = 1',' Lnk4 = 1',' Lnk5 = 1',' Frm1 =',' Begin Frame',' Naxes = 3',' Domain = "GRID"',' Ax1 ='... ... ' End Sp...',' MapB =',' Begin ZoomMap',' Nin = 1',' IsA Mapping',' Zoom = 0.001',' End ZoomMap',' End CmpMap',' End CmpMap',' End CmpMap',' End CmpMap',' ... End of Trace.
Task: plot the scan pattern of a SCUBA-2 map
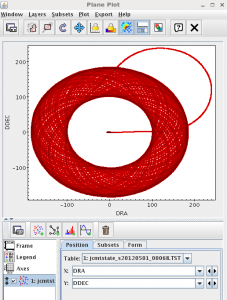
Use SMURF’s jcmtstate2cat to JCMT information raw SCUBA-2 observation (if you don’t have any raw SCUBA-2 observations on disk, an example output catalogue is provided in the file jcmtstate_s20120501_00068.TST in this tutorial’s tarball.). Use TOPCAT to plot the scan pattern.
Show worked example on clickYou can use all the same tools listed above to look at raw data as well. However, there is another tool that is useful for raw data: jcmtstate2cat in SMURF. This lets you see the JCMTSTATE extension in a raw file which contains timestreams of information about the position of the telescope, the instrumental data and the WVM. To avoid packaging up all the raw files, we’ve included the output catalogue here. If you needed to generate the file, here we show how to do it on from the s8a sub arrays of one of the observations in the SCUBA-2 basic DR tutorial.
smurf jcmtstate2cat ../SCUBA2_tutorial/raw/s8a20120501_00068*.sdf > jcmtstate_s20120501_00068.TST
This stores the catalogue in a file named jcmtstate_s20120501_00068.TST. You can open and plot this in Topcat.
topcat -f TST jcmtstate_s20120501_00068.TST&
This opens the file in TOPCAT, and tells TOPCAT that the file is in TST format.
Try creating a plot of the telescope scan position. You would normally do this by going to the Scatter Plot button, then selecting the DRA and DDec columns to plot. Try making other plots!
Measuring Noise in the Map
Task: Comparing noise estimates
Compare the noise estimated in a blank region of the spectra with the noise found in the Variance array
Show worked example on click
Worked Example
Take a reduced spectral cube, measure the noise on a blank part of the spectrum. Compare this noise with the measured noise from the Variance array.
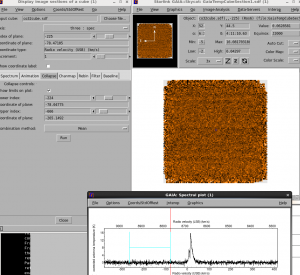
First of all, we will open our cube in Gaia in order to identify a blank region of the spectra over which to measure the noise.
The region from -265.0 to -78 km/s looks like it is very blank — we will use this to measure the noise.
You can either perform the collapse within Gaia by using the ‘collapse’ button on the Spectral cube window, and then either save the file or use the imaging tool in Gaia to examine the statistics, or you can use the command line. The command line is more repeatable.
First we will use KAPPA’s collapse command to collapse over the blank region. This command supports a wide range of estimators allowing you to produce integrated maps, RMS estiamtes and moment maps. Please see the documentation for full details. Here we will use the sigma estimator to get the standard deviation in that region.
collapse in=co32_cube.sdf axis=vrad low=-265.0 high=-78.0 estimator=sigma out=co32_noisemap.sdf
Note that we have to give the velocity range as floating points; if we give integers it will assume we mean pixel coordinates instead of km/s.
Now use the stats command to look at the values.
$ stats co32_noisemap.sdf
Pixel statistics for the NDF structure
/export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/co
32_noisemap
Title : Ros6
NDF array analysed : DATA
Pixel sum : 11429.2
Pixel mean : 0.829648
Standard deviation : 0.390908
Skewness : 2.14229
Kurtosis : 4.21748
Minimum pixel value : 0.518394
At pixel : (-12, -13)
Co-ordinate : (6:34:43.1, 4:11:20)
Maximum pixel value : 2.65666
At pixel : (60, -30)
Co-ordinate : (6:34:14.2, 4:09:38)
Total number of pixels : 14042
Number of pixels used : 13776 (98.1%)
No. of pixels excluded : 266 (1.9%)
Across the whole map we have a noise of 0.83+/-0.39 K
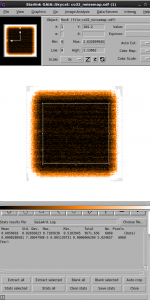
You can also open this map in Gaia, and use the Image Analysis->Image regions toolbar to measure the noise only in the center of the map.
gaiadisp co32_noisemap.sdf
Task: Regrid map and check the noise
Regrid the map into 1 km/s channels and see how the noise changes compared with before.
Show Worked solution on clickWorked Example
We will use sqorst (SQuash OR STretch) to rebin the data onto 1.0 km/s channels. We will accept the default method of rebinning.
sqorst in=co32_cube.sdf axis=3 mode=pixelscale pixscale=1.0 out=co32_1kms.sdf method=auto
Now, we will repeat the previous collapse command on this file in order to measure the noise after rebinning. Because we set the collapse limits in terms of a velocity instead of using a pixel index, we don’t have to change them after rebinning.
collapse in=co32_1kms.sdf axis=vrad low=-265.15 high=-78.05 estimator=sigma out=co32_1kms_noisemap.sdf
As before, we see the results with stats:
$ stats co32_1kms_noisemap.sdf
Pixel statistics for the NDF structure
/export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/co
32_1kms_noisemap
Title : Ros6
NDF array analysed : DATA
Pixel sum : 8072.22
Pixel mean : 0.585963
Standard deviation : 0.277194
Skewness : 2.13845
Kurtosis : 4.24379
Minimum pixel value : 0.341995
At pixel : (17, -12)
Co-ordinate : (6:34:31.5, 4:11:26)
Maximum pixel value : 1.91068
At pixel : (-19, -58)
Co-ordinate : (6:34:45.9, 4:06:50)
Total number of pixels : 14042
Number of pixels used : 13776 (98.1%)
No. of pixels excluded : 266 (1.9%)
Here you can see we now have a mean RMS noise in our regridded map of 0.59+/0.28 K. if you wish, you can verify that this has approximately followed the radiometer equation.
We can also compare the noise we have measured on the map with the noise in the VARIANCE component of the file. The easiest way is using the ‘comp’ option in stats to set which component to analyse (DATA is the default), and selecting the ERROR component, defined as the square root of the variance component.
$stats co32_1kms.sdf comp=ERROR
Pixel statistics for the NDF structure
/export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/co
32_1kms
Title : Ros6
NDF array analysed : ERROR
Pixel sum : 6.0305e+06
Pixel mean : 0.550634
Standard deviation : 0.267865
Skewness : 2.21899
Kurtosis : -2.97147
Minimum pixel value : 0.364528
At pixel : (-37, 6, -397)
Co-ordinate : (6:34:53.1, 4:13:14, -380.4388)
Maximum pixel value : 1.63818
At pixel : (57, 51, -397)
Co-ordinate : (6:34:15.4, 4:17:44, -380.4388)
Total number of pixels : 11163390
Number of pixels used : 10951920 (98.1%)
No. of pixels excluded : 211470 (1.9%)
We see a noise from the variance component of 0.55 +/- 0.26 K, compared with 0.59 +/- 0.28 K measured across one part of the empty spectra. The variance component was generated by SMURF’s gridding command ‘makecube’, and be default is generated from the Tsys values of the input spectra.
Task: measure the noise in a map
Measure the noise across a SCUBA-2 map. Use the scuba2_map.sdf file provided with this tutorial.
Worked solutionWorked Example
We will use the SCUBA-2 map provided in the sample data, <code>scuba2_map.sdf</code>. This is the same as the <code>gs20120501_68_850_reduced.sdf</code> file provided in the SCUBA-2 tutorial <code>example_reduced</code> data directory
There are several ways you can do this. The simplest is to examine the ERROR component of the data.
$stats scuba2_map.sdf comp=ERROR
Pixel statistics for the NDF structure
/export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/sc
uba2_map
Title : G34.3
NDF array analysed : ERROR
Pixel sum : 1640087.30197531
Pixel mean : 41.0555547705848
Standard deviation : 50.659757537598
Skewness : 3.90590482923681
Kurtosis : 26.0299464638736
Minimum pixel value : 10.6386591437756
At pixel : (-3, 23, 1)
Co-ordinate : (18:53:19.667, 1:16:26.30, 0.00085)
Maximum pixel value : 1157.3041341558
At pixel : (-13, 122, 1)
Co-ordinate : (18:53:22.334, 1:23:02.30, 0.00085)
Total number of pixels : 58081
Number of pixels used : 39948 (68.8%)
No. of pixels excluded : 18133 (31.2%)
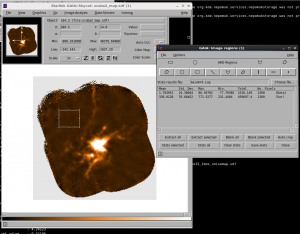
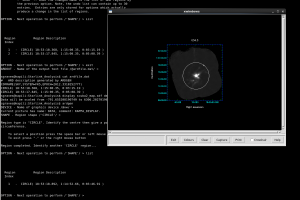
You can also see this in Gaia by going to Image Analysis-> Image Regions. Select the region you want to get statistics over and use the ‘Stats Selected’ button to generate the statistics. you can also use this to compare the noise in a blank part of the sky with the noise in the variance component.
In the example shown in the screen shot above, the Std. Deviation of the data area is 24.4 (measuring the RMS noise in that region); by comparison the noise in the Variance array has a mean value in that region of 338, corresponding to an error of 18.4. Fairly close, but there is clearly some difference!
Another method, designed to be used on reductions of a single observation, is to run the PICARD recipe SCUBA2_MAPSTATS on your file. This would like so:
$ picard -log sf -nodisplay SCUBA2_MAPSTATS scuba2_map.sdf Picard Says: No display will be used Picard Says: Pre-starting mandatory monoliths...Done Checking for next data file: /export/data/sgraves/sd/tutorial-data/JCMT_HETERODYNE_analysis_tutorial_2016/scuba2_map.sdf Storing: scuba2_map Picard Says: Creating temporary bad observation rules file A new group 20120501#-1 has been created Overriding PICARD instrument class to PICARD_SCUBA2_850 Sorting Groups REDUCING: scuba2_map Using recipe SCUBA2_MAPSTATS specified on command-line Processing data for G34.3 Calling _WRITE_MAPSTATS_LOGFILE_: write logfile with results from image analysis Writing results to log.mapstats... Cropping map using: RA= 18:53:18.6, Dec= 01:14:58.26, RADIUS=120. RMS is 13.5415200736753 Recipe took 2.018 seconds to evaluate and execute. Picard processing complete Processed one recipe which completed successfully Exiting...
In the output directory, you will now find a logfile anmed log.mapstats:
$cat log.mapstats
# Map results: noise, NEFD, exp_time log file - created on Thu Jan 25 20:45:30 2018 UT
#
# (YYYYMMDD.frac) (YYYY-MM-DDThh:mm:ss) () () () (um) (deg) () () () () (s) (s) () () () () (") (") () () () () ()
# UT HST Obs Source Mode Filter El Airmass Trans Tau225 Tau t_elapsed t_exp rms rms_units nefd nefd_units RA Dec mapsize pixscale project recipe filename
20120501.6984144 2012-05-01T06:45:43 68 G34.3 daisy 850 52 1.26 0.783 0.047 0.194 318 68.08 1.354E+01 mJy/bm 1.078E+02 mJy.s^0.5 18:53:18.6 01:14:58.26 240.0 4.0 M12AEC05 REDUCE_SCAN scuba2_map
This summarises information about the observation, as well as calculating the noise over central portion of the MAP, and the NEFD (Noise Equivalent Flux Density, usually used for tracking instrumental performance). If you run this command on a coadded map, it cannot accurately fill in most of the columns about the observation.
Masking, Extracting and Trimming
Task: Identifying the NDF pixel coordinates of a region with Gaia
Show worked solutionWorked Example
By default, the X: and Y: boxes in Gaia will show you the pixels relative to the bottom left of the map. Normally, the NDF PIXEL coordinates (relative to the pixel origin shown in ndftrace) are more useful. You can see these values in Gaia in two ways: one, you can open Image Analysis->Change Coordinates-> Show All Coordinates. This will bring up a seperate window that will show you the PIXEl (as well as GRID, AXIS< FRACTION and SKY) coordinates of your cursor as you move it around in the Gaia window.
Alternatively, you can start Gaia with the -pixel_indices=1 flag, and it will then show you the NDF PIXEL coordinates by default.
gaia -pixel_indices 1 scuba2_map.sdf
When you need to identify the pixel ranges, you can then use these coordinates. Test this out by finding the pixel values for a section of the map, then use ndfcopy and an NDF section to copy that part out. Open your new map in Gaia and verify you copied the part you were looking for. Remember that pixel indices have to be given in integers!
ndfcopy scuba2_map.sdf\(10:65,-5:53\) extracted.sdf gaia -pixel_indices 1 extracted.sdf &
Task: extract a region using WCS coordinates.
Try and extract the region from RA/Dec diagonal corners:(18:53:16, 1:14:35) to (18:53:01, 1:18:27.63)
Show worked solutionWorked Example
The main difficulity with this is formatting the strings correctly and using shell escapes. D There are several ways this could be done. One example is
ndfcopy scuba2_map.sdf\(18h53m16.3s:18h53m01.4s,1d14m35s:1d18m27.6s\) extracted2.sdf
In this example, don’t include any spaces next to the comma in the centre of the NDF section or it won’t be parsed correctly. If instead of using backslashes for your shell escapes you enclose the entire NDF and NDF section inside double and single quotes, then you can use spaces after the comman. E.g.:
ndftrace "'scuba2_map.sdf(18h53m16.3s:18h53m01.4s, 1d14m35s:1d18m27.6s)'"
NDF sections can be passed to commands anywhere an entire NDF could be passed. Once you get the hang of them they are a very useful feature of Starlink.
Task: CROP a SCUBA-2 image with PICARD
Use PICARD to crop a SCUBA-2 image into the requested area of the original observation with PICARD. An advanced option: try varying the radius.
Show worked solutionWorked Example
You can do this using the PICARD task CROP_SCUBA2_IMAGES:
$ picard -log sf CROP_SCUBA2_IMAGES scuba2_map.sdf Setting up display infrastructure (display tools will not be started until necessary)...Done Picard Says: Pre-starting mandatory monoliths...Done Checking for next data file: /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/scuba2_map.sdf Storing: scuba2_map A new group 20120501#-1 has been created Overriding PICARD instrument class to PICARD_SCUBA2_850 Sorting Groups REDUCING: scuba2_map Using recipe CROP_SCUBA2_IMAGES specified on command-line Processing data for G34.3 Calling _CROP_SCUBA2_IMAGE_: trim image to specified map size Trimming image to specified map size Trimming scuba2_map... Cropping map using: RA= 18:53:18.6, Dec= 01:14:58.26, RADIUS=120. Removing temporary files... Checking scuba2_map_crop... Keeping extension Recipe took 1.308 seconds to evaluate and execute. Picard processing complete Processed one recipe which completed successfully Exiting... Picard Says: Goodbye
By default this will use the center position of the original observation, and the ‘radius’ of the central area of the SCAN type, and produce a square cut out around the center.
If you wish to crop out a circular area or a larger area, you can provide a recpars file to alter this. E.g. to crop out a CIRCLE of radius 250″, you would save in a text file:
[CROP_SCUBA2_IMAGES] CROP_METHOD=CIRCLE MAP_RADIUS=250
Save this file as e.g. recpars.txt and then run
$ picard -log sf CROP_SCUBA2_IMAGES -recpars=recpars.txt scuba2_map.sdf Setting up display infrastructure (display tools will not be started until necessary)...Done Picard Says: Pre-starting mandatory monoliths...Done Checking for next data file: /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/scuba2_map.sdf Storing: scuba2_map A new group 20120501#-1 has been created Overriding PICARD instrument class to PICARD_SCUBA2_850 Sorting Groups REDUCING: scuba2_map Using recipe CROP_SCUBA2_IMAGES specified on command-line Processing data for G34.3 Calling _CROP_SCUBA2_IMAGE_: trim image to specified map size Trimming image to specified map size Output image will be a circle of radius 250 arcsec Masking the weights and exposure time images... Removing temporary files... Checking scuba2_map_tmpmask... Removing Checking scuba2_map_crop... Keeping extension Recipe took 2.668 seconds to evaluate and execute. Picard processing complete Processed one recipe which completed successfully Exiting... Picard Says: Goodbye
Task: ARD Masking
Make an ARD file and use the ardmask command to extract out an annular region around a point you choose.
ARD docs: http://starlink.eao.hawaii.edu/docs/sun183.htx/sun183.html
KAPPA guide to using ARD:
http://www.starlink.ac.uk/cgi-bin/htxserver/sun95.htx/sun95.html/?xref_se_ardwork
ARD format files are fairly straight forward, so they can be made yourself. A simple file to extract out a ring of inner radius 70″, outer radius 100″ and centered on RA=18:53:18.7, Dec=1:14:54.3 would be:
COFRAME(SKY, SYSTEM=FK5, equinox=2000) CIRCLE(18:53:18.7,1:14:54.3, :1:40.) .AND. .NOT. CIRCLE(18:53:18.7,1:14:54.3, :1:10.0)
Note that we define the radii in(third element of the CIRCLE command) in degrees:arcminutes:arcseconds. To create an annulus we have defined one larger circle, and then said to exclude the central region of the circle.
We save this as e.g. file ARD_ring.ard, and then we can apply it with ardmask.
ardmask in=scuba2_map.sdf ardfile=ARD_ring.ard out=scuba2_map_2d.sdf inside=False
Note that we need to remove the redundant third axis of the data — here we do this with ndf copy.
ndfcopy in=scuba2_map.sdf out=scuba2_map2d.sdf trim=true trimwcs=true
The result should look like:
You could also try using the ARDGEN interactive utility to create an ARD file. First of all, you need to display the image in an Xwindow using the KAPPA’s display command:
display scuba2_map.sdf dev=xw mode=faint
(This displays the map in the xwindows graphics device, using mode=faint so that you are not prompted to give a low and high value to scale between.)
You then run the command ardgen dev=xw and follow the prompts, selecting the shape you want to make and clicking in the xwindow to mark shapes. Remember you can use the ? at a command line prompt to see the help.
Task: masking based on SNR
- Mask out all pixels in a cube or map with a detection of less than 5 sigma (try errclip).
- Mask out all spectra in a cube where the peak of the spectra has a detection of less than 5 sigma. Try using makesnr, thresh, manic and copybad.
Mask out all pixels in a cube or map with a detection of less than 5 sigma (try errclip)
The command errclip can be used to clip a map or cube based on SNR levels, using the variance component of the map to define the snr. You can use it like:
errclip in=co32_cube.sdf mode=SNR out=co32_clipped5sigma limit=5.0
Open the resulting map in Gaia and take a look at it: you’ll see that the baseline and linewing regions are marked as bad, and only the strong parts of the line centre in bright regions are remaining.
Mask out all spectra in a cube where the peak of the spectra has a detection of less than 5 sigma. Try using makesnr, thresh, manic and copybad.
Often, its useful not to clip out individual channels on the basis of the noise limit, but instead to clip out only whole spectra where the peak was not detected at greater than 5 sigma.
First of all, we will need to collapse the cube down to a map of the peak values in each spectrum, using collapse. You can determine the velocity range to perform the collapse over by inspecting the cube in Gaia.
collapse in=co32_cube.sdf axis=vrad low=-6.5 high=25.67 estimator=max out=maxvalue
(If you would prefer to clip based on a detection in the integrated line, you could use the estimator=integ option instead.)
We will now create an SNR map from this 2-D peak map with the makesnr command:
makesnr in=maxvalue.sdf out=snr.sdf minvar=0.0
Then we use the thresh command to set bad all pixels with an SNR less than 5. We also select a spurious high value to set bad, as we have to provide both thrlo and thrhi options; this won’t cause any problems.
thresh in=snr out=snr_thr5 thrlo=5.0 thrhi=5000 newlo=bad newhi=bad
You now need to grow this mask into a 3-D mask you can apply to your original map. To do this we first need to find out what the pixel indices of are input cube are;
$ ndftrace co32_cube.sdf
NDF structure
/export/data/sgraves/sd/tutorial-data/Starlink_Analysis/co32_cube:
Title: Ros6
Label: TA* corrected antenna temperature
Units: K
Shape:
No. of dimensions: 3
Dimension size(s): 119 x 118 x 1880
Pixel bounds : -57:61, -58:59, -940:939
Total pixels : 26398960
Data Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
Variance Component:
Type : _REAL
Storage form: SIMPLE
Bad pixels may be present
World Co-ordinate Systems:
Number of co-ordinate Frames: 5
Current co-ordinate Frame (Frame 5):
Frame title : "3-d compound coordinate system"
Domain : SKY-DSBSPECTRUM
First pixel centre : 6:35:01.2, 4:06:50, -381.1352
Axis 1:
Label : Right ascension
Units : hh:mm:ss.s
Nominal Pixel scale: 5.99999 arc-sec
Axis 2:
Label : Declination
Units : ddd:mm:ss
Nominal Pixel scale: 5.99999 arc-sec
Axis 3:
Label : Radio velocity (USB)
Units : km/s
Nominal Pixel scale: 0.4233065 km/s
Extensions:
FITS
SMURF
PROVENANCE
History Component:
Created : 2014 Nov 20 09:22:05
No. records: 22
Last update: 2018 Jan 25 22:26:20 (WCSATTRIB (KAPPA 2.5-5))
Update mode: NORMAL
We can see here that our c32_cube goes from -940:939 in pixels in the 3rd, velocity, axis. We will now use the command manic to grow our 2D mask into a 3D mask we can apply to our original data; manic increases the dimensionality of the cube by (in this case) repeating the image plane pixels in every plane we add in the third dimension. The command we use is:
manic in=snr_thr5 out=cube_mask axes=\[1,2,0\] ubound=939 lbound=-940
Finally, we copy the bad pixel mask from this cube to our original CO 3-2 cube, and then open the masked map in Gaia to examine it.
copybad in=co32_cube.sdf ref=cube_mask.sdf out=co32_masked.sdf gaiadisp co32_masked.sdf
Extracting Values
You can always of course use the stats command to get the sum/mean etc value for small regions, or within extracted NDF sections or ARD masked/extracted areas, and you may find that sufficient for reading out some of your values. The pluck command can be used to easily get data values at a specific point on the map however, or to extract the whole spectra from that point.
Task: Using pluck to extract spectra
Using the co32_cube.sdf provided with this tutorial, extract spectra from the following coordinates:
6:34:37.7,4:12:37.20, 6:34;32.77,4:13:09.77, 6:34:24.47, 4:08:56.91 . Examine the output spectra, either with Splat or by using linplot
You can use pluck interactively, but here we will use it by giving it a text file. The format of the input catalogue is:
6:34:37.7 4:12:37.20 1 6:34:32.77 4:13:09.77 1 6:34:24.47 4:08:56.91 1
Note that you have to provide a value for axis 3, even though the output will include all values from that axis. We then run the command with:
$ pluck in=co32_cube.sdf axes=vrad mode=file coin=spectra_positions.txt container=True out=container_spectra.sdf method=linear Creating /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/container_spectra.PLUCK _1. Creating /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/container_spectra.PLUCK _2. Creating /export/data/sgraves/sd/tutorial-data/Starlink_Analysis/container_spectra.PLUCK _3.
We used a linear resampling method to extract the pixels; others are available, but may require setting the params paramter to use. See the documentation for details. We now have an output file container_spectra.sdf which contains 3 input spectra as separate NDFs. We can copy them out to their own files with:
ndfcopy container_spectra.PLUCK_1 pluck_1 ndfcopy container_spectra.PLUCK_2 pluck_2 ndfcopy container_spectra.PLUCK_3 pluck_3
If you use ndftrace to look at one of the spectra, it includes all the information about the spatial position it was extracted from.
$ ndftrace pluck_*.sdf
NDF structure
/export/data/sgraves/sd/tutorial-data/Starlink_Analysis/pluck_1:
Title: Ros6
Label: TA* corrected antenna temperature
Units: K
Shape:
No. of dimensions: 3
Dimension size(s): 1 x 1 x 1880
Pixel bounds : 1:1, 1:1, -940:939
Total pixels : 1880
Data Component:
Type : _REAL
Storage form: SIMPLE
There are no bad pixels present
Variance Component:
Type : _REAL
Storage form: SIMPLE
There are no bad pixels present
World Co-ordinate Systems:
Number of co-ordinate Frames: 5
Current co-ordinate Frame (Frame 5):
Frame title : "3-d compound coordinate system"
Domain : SKY-DSBSPECTRUM
First pixel centre : 6:34:37.7, 4:12:37, -381.1352
Axis 1:
Label : Right ascension
Units : hh:mm:ss.s
Nominal Pixel scale: 6.00000 arc-sec
Axis 2:
Label : Declination
Units : ddd:mm:ss
Nominal Pixel scale: 6.00000 arc-sec
Axis 3:
Label : Radio velocity (USB)
Units : km/s
Nominal Pixel scale: 0.4233065 km/s
Extensions:
FITS
SMURF
PROVENANCE
History Component:
Created : 2014 Nov 20 09:22:05
No. records: 24
Last update: 2018 Jan 26 02:41:53 (NDFCOPY (KAPPA 2.5-5))
Update mode: NORMAL
You can also view the spectra in splat with:
splat pluck_*.sdf&
Or display them with KAPPA’s linplot command:
linplot pluck_1 dev=xw
Coordinate conversion
Task: Convert a map to galactic coordinates
Use wcsattrib to change the Coordinate system of your CO 3-2 Ros6 map into Galactic Coordinates. Check the output map has the new coordinates
Show worked solutionWorked Example
Setting a new coordinate system in a map is easy. However, please note that this will not change the pixels in anyway: if you want to regrid your map onto a pixel grid aligned with a new coordinate system (this can matter more for extremely large maps), then you should look into the regrid command. Starlink NDF files, unlike FITS files, generally have multiple WCS systems associated with them, with defined transformations between them. Therefore, changing the coordinates only involves running wcsattrib to tell it to set the current system to your desired one. In this case, you will do:
wcsattrib ndf=co32_cube.sdf mode=set name=System newval=Galactic
And thats it!
You can verify it by opening and Gaia and seeing which coordinate system is the default, or by converting to a FITS file with:
convert ndf2fits co32_cube.sdf co32_galactic.fits
and opening it with your favourite non Gaia program. For example, see what the Python plotting program APLpy made of the FITS version of this file:
Task: Align one map with another.
This is already done as part of Tutorial 5: CO subtraction. Check there for the full details, or see the wcsalign documentation at: http://www.starlink.ac.uk/cgi-bin/htxserver/sun95.htx/sun95.html?xref_WCSALIGN
Fitting Spectra
Task: use fit1d to fit your entire cube
SMURF’s fit1d routine can be used to fit a 1D spectrum to every pixel in your cube. Try it out on the co32_cube.sdf or co32_masked.sdf.
Show worked solutionI highly recommend reading the documentation for fit1D: it has a lot of options, but is very powerful. As a quick overview, here is one example of using it.
First, create a configuration file. An example one can be found in $STARLINK_DIR/bin/smurf/smurf_fit1d.def , including descriptions of various parameters. We are going to use the basic paramters, and only provide updated values for the range parameter. This determines the radio velocity range we will fit the profiles within, and we will use -12 to 88 km/s.
This mostly-default config will fit a single Gausshermite2 to each spectrum. We define our config file as:
^$SMURF_DIR/smurf_fit1d.def range=(-12.0,88.0)
We save it as myfit1d.def, and then run our command as:
$ fit1d in=co32_masked.sdf out=fittedprofiles.sdf rms=! config=^myfit1d.def Copying input file 1 to output for fitting... Fitting data using 1 GAUSSHERMITE2(s) over pixel range [873,1109] ...Will use 16 threads to fit 14042 profiles. (FitProfileThread 16) ...at profile 14000 - 845 of 887 (i=26318120) ...Writing parameter cubes and finishing up.
(A Caret, ^, indicates to read the contents of the text file that follows.)
If you open the resulting cube up in Gaia, and also open the original map in Gaia next to it via the File->new Window option, you can use the Open Cube->Options->Slave to move about in one cube and see spectra from both cubes. Alternatively, you can use the sub command to view the residuals from the fit.
sub in1=co32_masked.sdf in2=fittedprofiles.sdf out=residuals.sdf
Clumpfinding
Task: Use fellwalker to find clumps.
Use the Fellwalker algorithm to find clumps of emission in the G34.3 SCUBA-2 map for this tutorial. Copy out some of the individual clumps from the CUPID extension and view them in Gaia.
Show worked solutionIf you’re not familiar with Fellwalker, you should read the description of the algorithm in the CUPID manual, or David Berry’s 2015 A&C paper.
We first of all need to select our configuration parameters. Here, we are going to look for all clumps of 5 sigma above the noise, and we will subdivide neighbouring clumps into separate clumps if they have gaps between their peaks of at least 3sigma. We will follow the clumps down to 3 sigma above the noise. The config file we need to define is then:
FellWalker.Noise=3*RMS FellWalker.MinHeight=5*RMS FellWalker.MinDip=3*RMS
We save that into a text file, here named config.lis, and then we can call findclumps (after initialising cupid) as:
findclumps scuba2_map.sdf out=scuba2_clumps.sdf \
outcat=clumpcat.FITS method=fellwalker deconv=no accept
We have to provide the special keyword accept indicating that all other parameters that would be prompted for should use their default value. This is necessary if you don’t want to be prompted to accept the default RMS value (calculated from the VARIANCE array);
findclumps will otherwise wait for you to either accept the calculated RMS or give a different value.
We now have an output catalog in clumpcat.FITS that we can open and view with Topcat, and an output clumpmask in scuba2_clumps.sdf that we can view in Gaia. Go ahead and open the mask in Gaia:
gaia scuba2_clumps.sdf&
You can see the output is a mask over the full area of each clump, where the value of that clump is the same as the clump index in the output catalogue. Pixels not assigned to a clump are set as BAD values.
The output file also has a special CUPID extension; for fellwalker, this contains an array of all the identified clumps with the data values from the original data included. You can access these like you do any other NDF extension, so to copy out (for example) clump 3 to its own NDF file you would do:
ndfcopy scuba2_clumps.sdf.more.CUPID.CLUMPS\(3\) clump3.sdf
Or you can even open that clump directly in Gaia with:
gaiadisp scuba2_clumps.sdf.more.CUPID.CLUMPS\(3\)